In vivo recognition of Bacillus subtilis by desorption electrospray ionizationmass spectrometry (DESI-MS)
Literature Information
Yishu Song, Nari Talaty, Kirill Datsenko, Barry L. Wanner, R. Graham Cooks
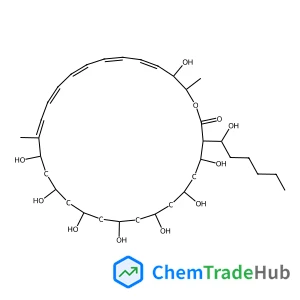
Desorption electrospray ionization mass spectrometry (DESI-MS) of culture of the bacterium Bacillus subtilis as a biofilm growing on agar nutrient gives simple, high quality mass spectra dominated in both the positive and negative ion modes by signals due to the cyclic lipopeptide, Surfactin(C15). This in vivo experiment, performed by direct analysis of untreated microorganism samples under ambient conditions, allows rapid identification of this microorganism and the antibiotics that it produces. The result is suggestive of the capabilities of DESI-MS for in vivo microorganism characterization in general and for monitoring fermentation processes for the production of antibiotics and other biochemicals.
Recommended Journals
Related Literature
IF 6.367
An overview of latest advances in exploring bioactive peptide hydrogels for neural tissue engineeringIF 6.843
Sensitive and specific detection of tumour cells based on a multivalent DNA nanocreeper and a multiplexed fluorescence supersandwichIF 6.222
Mechanically stable and economically viable polyvinyl alcohol-based membranes with sulfonated carbon nanotubes for proton exchange membrane fuel cellsIF 6.367
Synthesis and optical and electronic properties of one-dimensional sulfoxonium-based hybrid metal halide (CH3)3SOPbI3IF 6.222
A hollow neuronal carbon skeleton with ultrahigh pyridinic N content as a self-supporting potassium-ion battery anodeIF 6.367
Performance of electrode-supported silica membrane separators in lithium-ion batteriesIF 6.367
Small size yet big action: a simple sulfate anion templated a discrete 78-nuclearity silver sulfur nanocluster with a multishell structureIF 6.222
Co-production of pure hydrogen, carbon dioxide and nitrogen in a 10 kW fixed-bed chemical looping systemIF 6.367
Ether-functionalization of monoethanolamine (MEA) for reversible CO2 capture under solvent-free conditions with high-capacity and low-viscosityIF 6.367
Source Journal
Analyst

Analyst publishes analytical and bioanalytical research that reports premier fundamental discoveries and inventions, and the applications of those discoveries, unconfined by traditional discipline barriers.
Recommended Compounds
Recommended Suppliers
 Emile Egger & Cie SA
Emile Egger & Cie SA TKT-Kunststoff-Technik GmbH
TKT-Kunststoff-Technik GmbH Piller Industrieventilatoren GmbH
Piller Industrieventilatoren GmbH Nanjing Yanqiao Technology Co., Ltd.
Nanjing Yanqiao Technology Co., Ltd. Zhucheng Taisheng Chemical Co., Ltd.
Zhucheng Taisheng Chemical Co., Ltd. Hangzhou Changhe Chemical Industry Co., Ltd.
Hangzhou Changhe Chemical Industry Co., Ltd. Elektrowärme Gabbey
Elektrowärme Gabbey QUANCOM Informationssysteme GmbH
QUANCOM Informationssysteme GmbH Linde AG, Linde Engineering Division
Linde AG, Linde Engineering Division Elite Thermal Systems Ltd
Elite Thermal Systems Ltd