Nature Methods
Basic Information
Nature Methods is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences. This monthly publication is aimed at a broad, interdisciplinary audience of academic and industry researchers actively involved in laboratory practice. It provides researchers with new tools to conduct their research and places a strong emphasis on immediate practical relevance and potential to advance new biological applications. The journal publishes method developments (Articles, Brief Communications), performance comparisons of related, established methods (Analyses), and collections of tools or datasets of broad interest (Resources) as primary research papers. The journal also publishes overviews of recent technical and methodological developments (Reviews, Perspectives) and opinion pieces (Comments). We are actively seeking papers of relevance to the broad life sciences basic research community, including methods grounded in chemistry, physics and computer science. To enhance the practical relevance of each paper, the description of the method must be accompanied by strong validation, an application to an important biological question and results illustrating its performance in comparison to available approaches. Papers are selected for publication based on broad interest, thorough assessments of methodological performance and comprehensive technical descriptions that facilitate immediate application. Specific areas of interest include, but are not limited to: Methods for genome sequencing and structure profiling Single cell analysis methods, including sequencing, 'omics and multi-omics approaches Methods for spatial 'omics Methods for metagenome analysis Methods for genome engineering Techniques for the analysis and manipulation of gene expression, including epigenetics, gene targeting, transduction Methods for proteomics, including mass spectrometry, analysis of binding interactions and analysis of post-translational modifications Methods for systems biology and interactome profiling Methods for glycobiology Methods for metabolomics Structural biology techniques, including cryo-electron microscopy, cryo-electron tomography, and crystallography Methods for protein engineering Chemical biology techniques, including chemical labeling, methods for expanding the genetic code and chemogenetic tools Biophysical methods, including single molecule and lab-on-a-chip technologies Optical and non-optical imaging technologies, including microscopy, spectroscopy and in vivo imaging Probe design, including genetically encoded and organic dye-based probes and sensors, as well as labeling strategies Methods for cell culture and manipulation, including microbiology approaches Methods to culture, manipulate and study stem cells, organoids and assembloids Immunological techniques, including antigen discovery methods, repertoire discovery methods and methods for studying immune cell interactions Methods for the study of physiology and disease processes including cancer, tumor microenvironment, and pathophysiology Neuroscience methods, such as approaches for studying the brain at different scales, including optical microscopy and MRI, and methods to study animal behavior Methods involving model organisms and their manipulation Computational, statistical and machine learning methods for analysis, modeling and visualization of biological data
CiteScore
| Subject | Rank | Percentile |
|---|---|---|
Biochemistry, Genetics and Molecular BiologyBiochemistry |
1 / 438 | 99% |
Journal Statistics
Submission Information
Submission Website:
https://author-welcome.nature.com/guided-oa/41592Accepted Types:
Related Articles
An aptasensing platform for simultaneous detection of multiple analytes based on the amplification of exonuclease-catalyzed target recycling and DNA concatemers
Liping Jiang, Jingdong Peng, Ruo Yuan, Yaqin Chai, Yali Yuan, Lijuan Bai, Yan Wang
DOI: 10.1039/C3AN00757J
Integration of nanoporous membranes into microfluidic devices: electrokinetic bio-sample pre-concentration
Minseok Kim
DOI: 10.1039/C3AN00965C
An electrogenerated chemiluminescence sensor prepared with a graphene/multiwall carbon nanotube/gold nanocluster hybrid for the determination of phenolic compounds
Dehua Yuan, Shihong Chen, Ruo Yuan, Juanjuan Zhang, Wen Zhang
DOI: 10.1039/C3AN01031G
A colorimetric and fluorometric dual-modal supramolecular chemosensor and its application for HSA detection
Junfeng Xiang, Xiufeng Zhang, Hongbo Chen, Qianfan Yang, Qian Li, Aijiao Guan, Qian Shang, Yalin Tang, Guangzhi Xu
DOI: 10.1039/C3AN01929B
Generation of a chemical gradient across an array of 256 cell cultures in a single chip
Himali Somaweera, Akif Ibragimov
DOI: 10.1039/C3AN00946G
Dual signal amplification of zinc oxide nanoparticles and quantum dots-functionalized zinc oxide nanoparticles for highly sensitive electrochemiluminescence immunosensing
Jinxing Zhang, Suli Liu, Jianchun Bao, Wenwen Tu, Zhihui Dai
DOI: 10.1039/C3AN00705G
A compact high resolution ion mobility spectrometer for fast trace gas analysis
Ansgar T. Kirk, Maria Allers, Philipp Cochems, Jens Langejuergen, Stefan Zimmermann
DOI: 10.1039/C3AN00231D
Design of a universal biointerface for sensitive, selective, and multiplex detection of biomarkers using surface plasmon resonance imaging
Arghavan Shabani
DOI: 10.1039/C3AN01374J
Proteomics applied to the authentication of fish glue: application to a 17th century artwork sample
Sophie Dallongeville, Mark Richter, Stephan Schäfer, Michael Kühlenthal, Nicolas Garnier, Christian Rolando, Caroline Tokarski
DOI: 10.1039/C3AN00786C

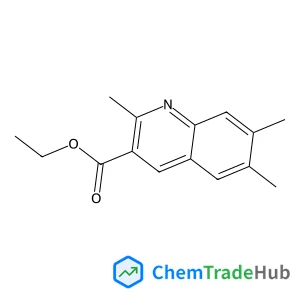
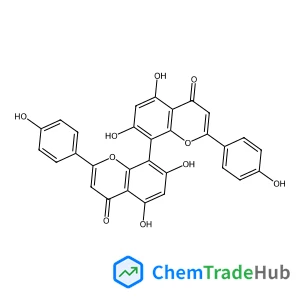
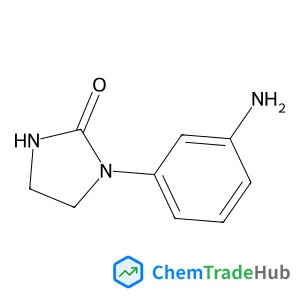
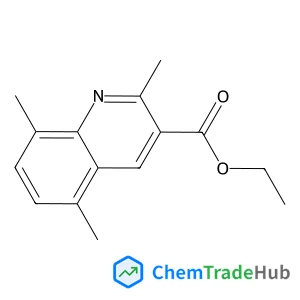
Recommended Compounds
Recommended Suppliers
 Pepperl+Fuchs SE
Pepperl+Fuchs SE Yangzhou Pharmaceutical Co., Ltd.
Yangzhou Pharmaceutical Co., Ltd. ita-Tech GmbH
ita-Tech GmbH Philipp Kirsch GmbH
Philipp Kirsch GmbH Wuhan Esitep Technology Co., Ltd.
Wuhan Esitep Technology Co., Ltd. Hangzhou Kenos Technology Biology Co., Ltd.
Hangzhou Kenos Technology Biology Co., Ltd. Lanson Laboratory Equipment Co., Ltd
Lanson Laboratory Equipment Co., Ltd INNOCISE GmbH
INNOCISE GmbH Riyuecao (Shandong) Biopharmaceutical Technology Co., Ltd.
Riyuecao (Shandong) Biopharmaceutical Technology Co., Ltd. Guangzhou Xitang Electromechanical Technology Co., Ltd.
Guangzhou Xitang Electromechanical Technology Co., Ltd.








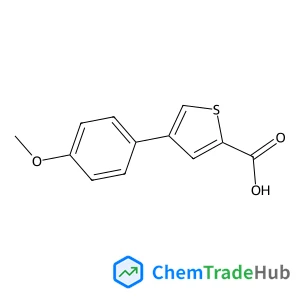
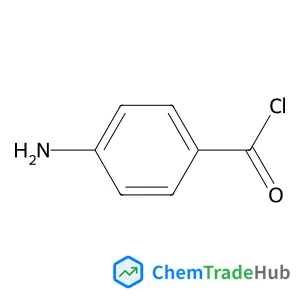
![175202-29-6 - 2-[1-Methyl-3-(trifluoromethyl)pyrazol-5-yl]thiophene-5-carboxylic acid 175202-29-6 - 2-[1-Methyl-3-(trifluoromethyl)pyrazol-5-yl]thiophene-5-carboxylic acid](/structs/175/175202-29-6-812a.webp)
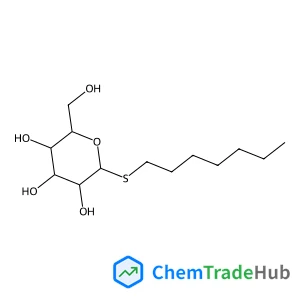
![2757-90-6 - (2S)-2-Amino-5-{2-[4-(hydroxymethyl)phenyl]hydrazino}-5-oxopentanoic acid (non-preferred name) 2757-90-6 - (2S)-2-Amino-5-{2-[4-(hydroxymethyl)phenyl]hydrazino}-5-oxopentanoic acid (non-preferred name)](/structs/275/2757-90-6-7436.webp)
![52450-38-1 - N-[3-(2-Formamido-5-methoxyphenyl)-3-oxopropyl]acetamide 52450-38-1 - N-[3-(2-Formamido-5-methoxyphenyl)-3-oxopropyl]acetamide](/structs/524/52450-38-1-ad06.webp)
![57322-42-6 - 3-[(E)-(4-Anilino-1-naphthyl)diazenyl]-2,7-naphthalenedisulfonic acid 57322-42-6 - 3-[(E)-(4-Anilino-1-naphthyl)diazenyl]-2,7-naphthalenedisulfonic acid](/structs/573/57322-42-6-b26c.webp)